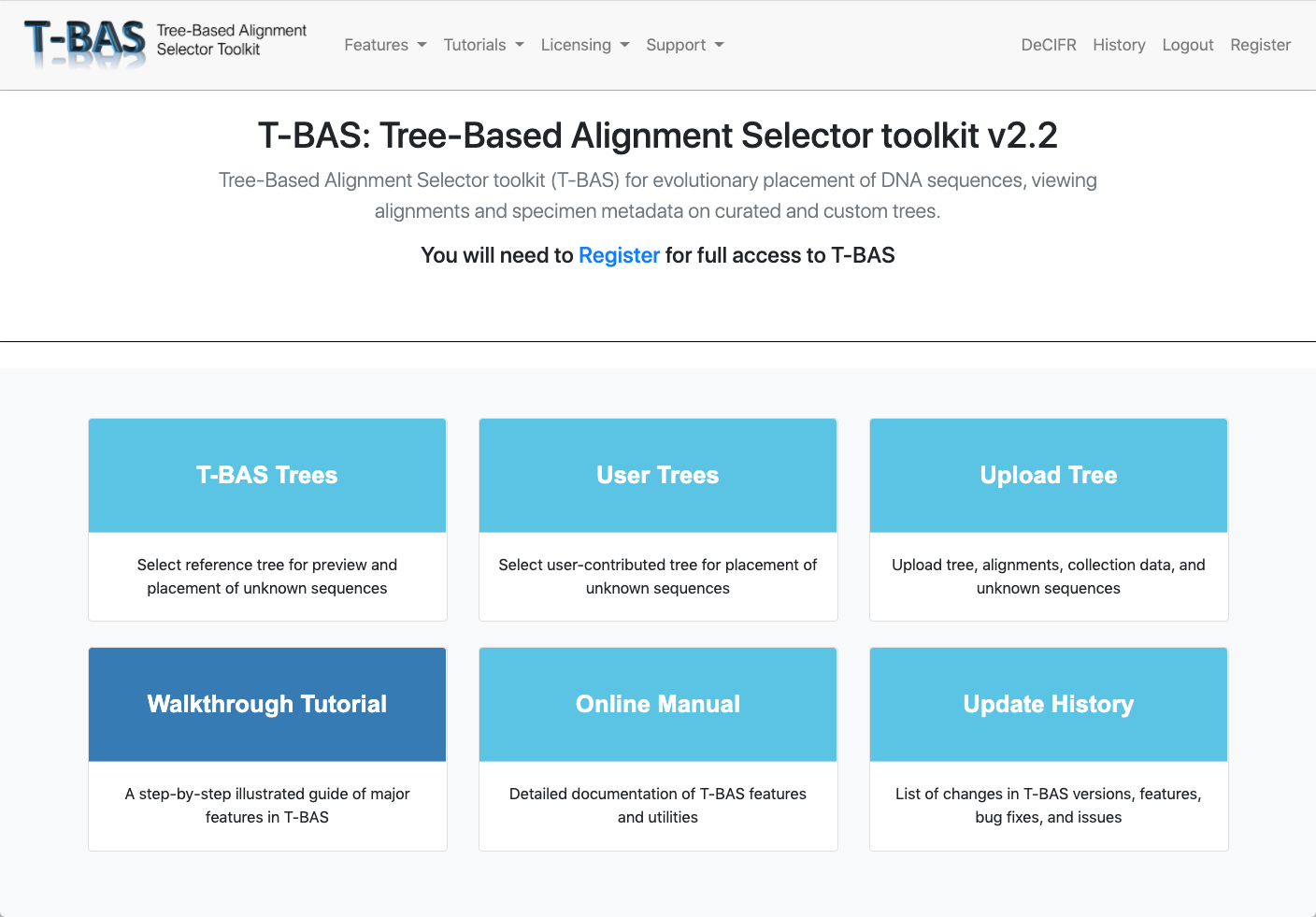
Reference and Custom Trees
Select from a diverse set of trees across all three domains of life (Archaea, Eukarya, Bacteria) and viruses or upload your own tree.
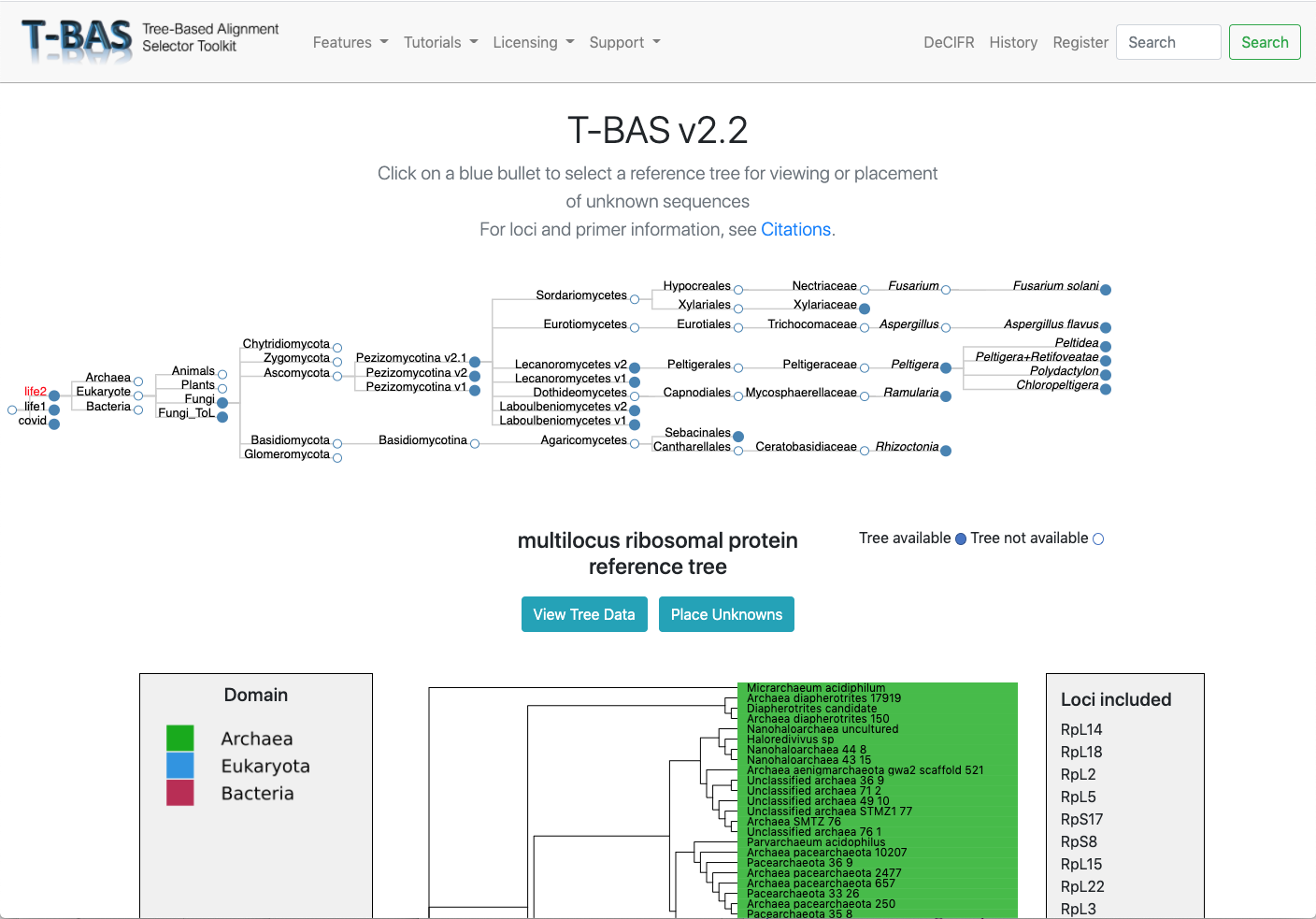
Multi-level Phylogenetic Placement
Place unknown sequences using single or multiple loci across trees from domain to species, and on the population level.
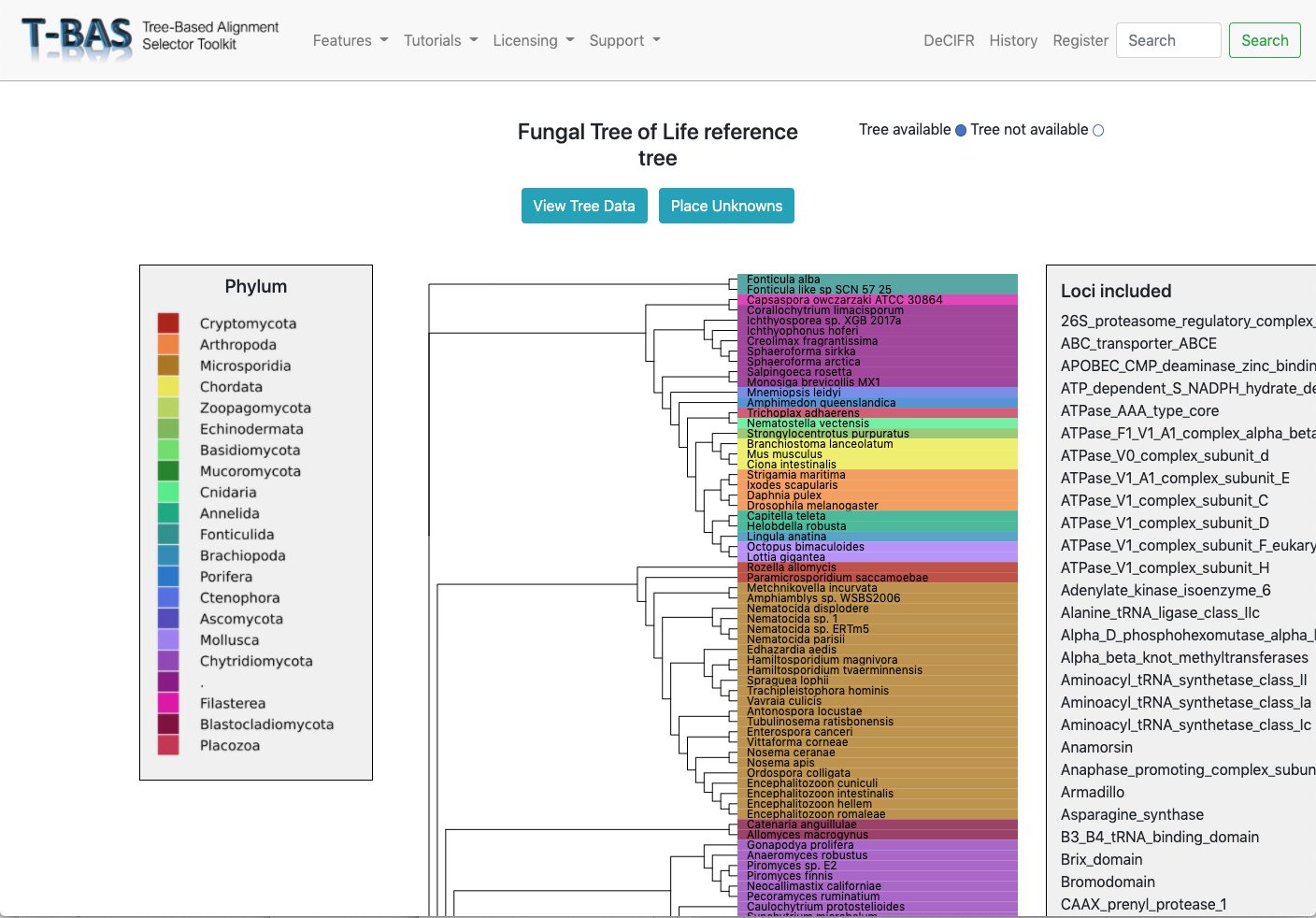
Genomes and Metagenomes
Place genomes and metagenomes on 16-locus domain level protein tree or 290-locus kingdom fungal tree.
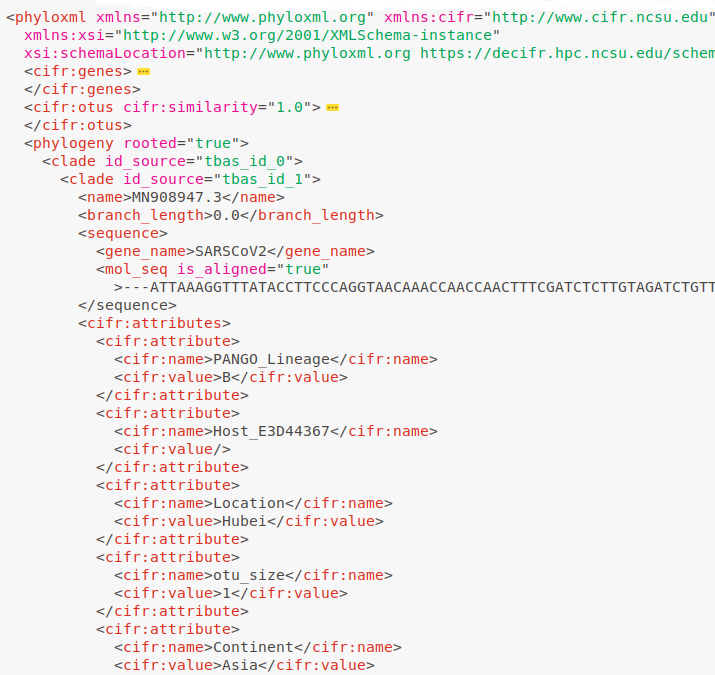
Data Standardization
Save trees, alignments and metadata in Metadata Enhanced PhyloXML (MEP) format, facilitating data validation and tracking.

Trees, Clades and Attributes
Reconstruct multi-locus trees and networks with increased taxon sampling for specimen attributes of interest.

Public and Docker Development
Explore tool features on public version or purchase a private license to use T-BAS within Docker.